Should You Get a Gut Microbiome Test? A Gastroenterologist’s Evidence-Based Perspective


Last week, a patient arrived in my clinic with results from a direct-to-consumer gut microbiome test. The report labeled her as “deficient” in multiple bacterial species and recommended several probiotic supplements. It included polished graphics, percentile rankings, and a composite “dysbiosis score.”
The challenge was not the presentation—it was the interpretation. The test measured relative bacterial abundance, not microbial function. The reported “deficiencies” reflected comparisons to a reference population of uncertain clinical relevance, rather than actionable biological deficits. Without functional context, these results risked driving unnecessary interventions and expense.
This scenario is increasingly common. As gut microbiome testing becomes more accessible, patients and clinicians alike are confronted with reports that appear precise but often lack clinical clarity. Understanding what these tests measure, their limitations, and when they add real value is essential.
In this article, I will clarify how gut microbiome tests work, what they can and cannot tell us, and how to determine whether testing is appropriate for your health goals.
Executive Summary
- The gut microbiome plays a central role in metabolism, inflammation, immune regulation, and overall healthspan
- Gut microbiome testing analyzes stool samples using DNA sequencing to assess microbial composition and functional capacity
- The two primary testing methods—16S rRNA sequencing and shotgun metagenomic sequencing—offer different levels of insight and clinical utility
- Microbiome tests do not diagnose disease and represent a snapshot in time, not a fixed biological state
- When interpreted appropriately, microbiome testing can enhance precision care; when used in isolation, it can be misleading

The Core Question: What Are We Actually Measuring?
Modern microbiome tests use DNA sequencing to identify microorganisms in your stool. But not all sequencing methods are created equal, and understanding the difference is critical to interpreting what your results actually mean.
16S rRNA Gene Sequencing: The Industry Standard
This is what most consumer tests and many clinical labs use. It works by targeting a specific bacterial gene—the 16S ribosomal RNA gene—that serves as a genetic fingerprint for bacterial identification.
What it does well:
- Identifies bacteria at the genus level (like identifying "Bifidobacterium" but not which specific species)
- Provides a snapshot of overall microbial diversity
- Relatively affordable and fast
- Uses well-established reference databases
Critical limitations:
- Measures relative abundance, not absolute numbers. If you have less Bacteroides, it might mean you actually have fewer Bacteroides, or it could just mean other bacteria increased. The test can't tell the difference.
- Limited species/strain identification. This matters because Lactobacillus rhamnosus GG has clinical evidence, but "Lactobacillus genus" tells us almost nothing actionable.
- No functional information. It's like having a list of kitchen ingredients but no idea what meal you can make. Knowing bacteria are present doesn't tell you what they're producing.
My clinical perspective: 16S sequencing gives you a compositional overview—useful for assessing severe dysbiosis or tracking major shifts after intervention, but often insufficient for precision treatment planning.
Shotgun Metagenomic Sequencing: The Comprehensive Approach
This method sequences all the DNA in your stool sample—not just bacterial genes, but everything present.
What it offers:
- Species and strain-level identification
- Detection of bacteria, archaea, fungi, and viruses
- Functional profiling—analysis of metabolic pathways and gene capacity
- Insight into antimicrobial resistance genes
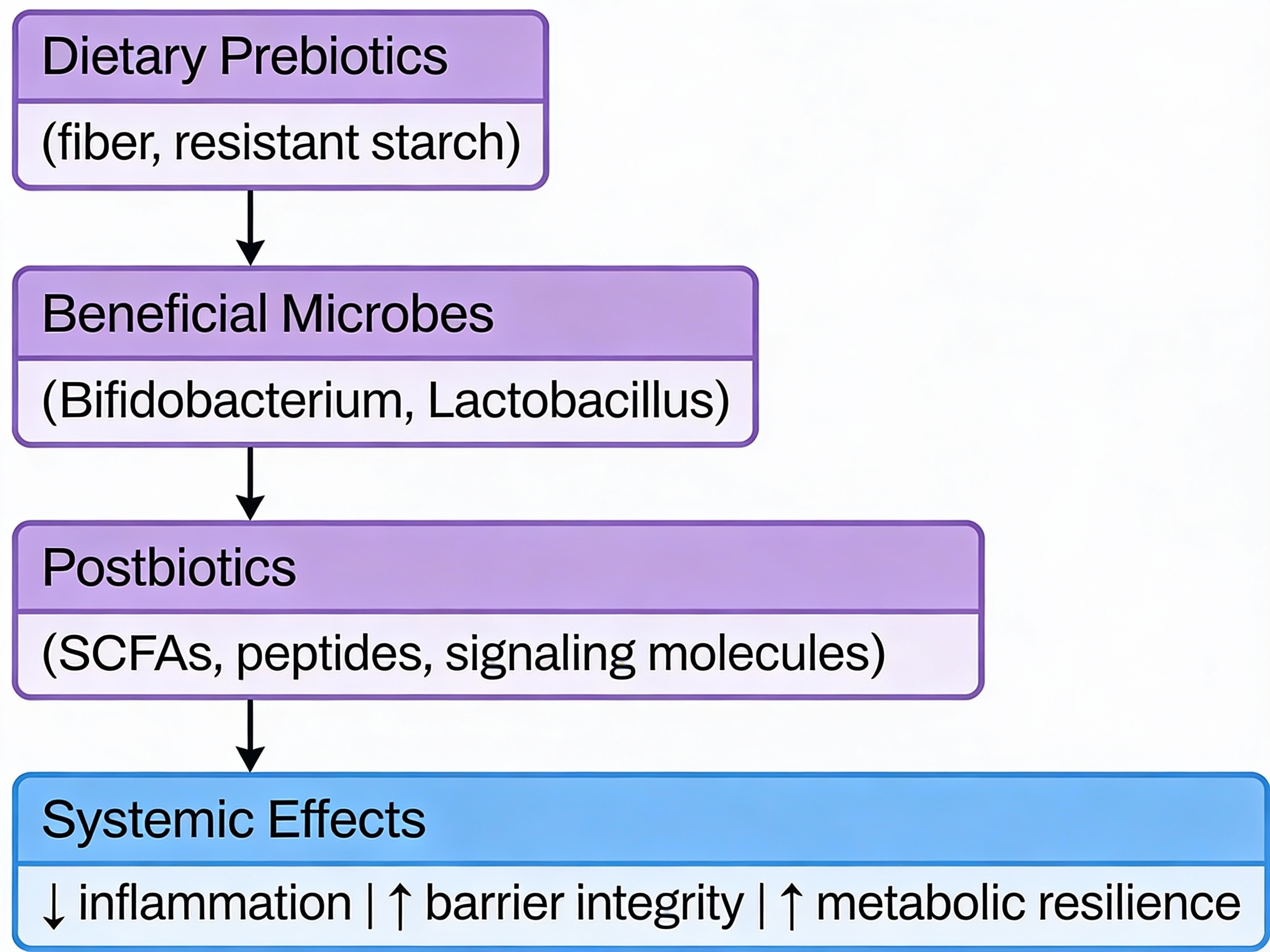
Why this matters for longevity: Functional data tells us whether your microbiome has the genetic machinery to produce butyrate, metabolize polyphenols, synthesize certain vitamins, or generate inflammatory compounds. This is clinically meaningful information that composition alone cannot provide.
The trade-offs:
- Higher cost
- More complex interpretation
- Requires sophisticated bioinformatics and clinical integration
- Still doesn't tell us what's actually being produced in real-time
When I order shotgun sequencing: For complex cases with treatment-resistant symptoms, metabolic dysfunction despite dietary optimization, or when I need to understand functional capacity before targeted interventions.
What Microbiome Testing Is Not
Let's clear up common confusion:
Traditional stool tests (culture, ova & parasite exams, PCR panels) are not microbiome tests. These look for specific pathogens—they're diagnostic tools for acute infections, not ecosystem profiling. They won't tell you about diversity, composition, or functional capacity.
Blood tests for "leaky gut" or food sensitivities are not microbiome tests. While gut barrier function and immune responses relate to the microbiome, these tests measure different phenomena and shouldn't be conflated with microbiome analysis.
The Technologies You'll Hear About Next
The field is evolving rapidly. Here's what's emerging from research into clinical practice:
Metatranscriptomics: Instead of DNA, this measures RNA. Think of it as the difference between having a recipe book and actually cooking the meal.
Metabolomics: Directly measures the chemical compounds produced by your microbiome—short-chain fatty acids, bile acids, tryptophan metabolites. This is the closest we get to understanding actual physiological impact, and in my view, represents the future of actionable testing.
Mycobiome and virome analysis: Specialized sequencing for fungal communities and bacteriophages. Emerging evidence suggests these play significant roles in health, but clinical applications are still being defined.
These technologies are mostly research-based today but will likely become standard within 5-10 years as costs decrease and interpretation frameworks mature.
My Clinical Framework
When patients ask about microbiome testing, I walk through these questions to maximize the value of their investment:
- What specific question are we trying to answer? "I want to optimize my gut health" is too vague. "I want to understand why I'm still bloated despite FODMAP elimination" is actionable and guides us toward the right test.
- Will the results guide personalized intervention? Testing shines when it reveals specific imbalances that direct targeted treatment—whether that's particular prebiotic fibers, antimicrobial protocols, or dietary modifications you wouldn't have tried otherwise.
- Are we measuring the right thing? Matching the test to your clinical picture maximizes insight. Functional analysis for metabolic concerns, compositional assessment for dysbiosis, targeted sequencing when specific organisms are suspected.
- How will we integrate results with your complete picture? Microbiome data becomes powerful when combined with your symptoms, dietary patterns, medication history, and inflammatory markers—this integration reveals patterns single data points miss.
- What's the intervention and reassessment plan? The greatest value comes from testing, implementing targeted changes, then retesting to confirm improvement—creating a feedback loop that refines your personalized protocol.
The Bottom Line
Microbiome testing can be a valuable clinical tool when:
- Ordered for specific indications, not general curiosity
- Interpreted by someone who understands both the technology and the clinical context
- Used to guide targeted interventions, not generate anxiety about bacterial "deficiencies"
- Followed by appropriate reassessment to measure intervention success.
The microbiome matters immensely for healthspan and longevity. But understanding what to measure, when to measure it, and how to act on results separates useful clinical data from expensive noise.
Dr. Banerjee is a board-certified gastroenterologist with over 15 years of clinical experience, peer-reviewed publications indexed in PubMed, and deep expertise in gut microbiome science. He advises high-achieving individuals and families on precision longevity and healthspan optimization. Expanded clinical analysis is available through the Private Longevity Briefing.
Selected Peer-reviewed evidence
- Chen S, Chen W, Wang X, Liu S. Mendelian randomization analyses support causal relationships between gut microbiome and longevity. J Transl Med. 2024;22(1):1032. Published 2024 Nov 16. doi:10.1186/s12967-024-05823-2
- Zhang G, Lu Y, Wang Z, et al. Causal relationship between gut microbiota and ageing: A multi-omics Mendelian randomization study. Arch Gerontol Geriatr. 2025;131:105765. doi:10.1016/j.archger.2025.105765
- Liu X, Zou L, Nie C, et al. Mendelian randomization analyses reveal causal relationships between the human microbiome and longevity. Sci Rep. 2023;13(1):5127. Published 2023 Mar 29. doi:10.1038/s41598-023-31115-8
- He D, Liu L, Zhang Z, et al. Association between gut microbiota and longevity: a genetic correlation and mendelian randomization study. BMC Microbiol. 2022;22(1):302. Published 2022 Dec 13. doi:10.1186/s12866-022-02703-x
- Porcari S, Mullish BH, Asnicar F, et al. International consensus statement on microbiome testing in clinical practice. Lancet Gastroenterol Hepatol. 2025;10(2):154-167. doi:10.1016/S2468-1253(24)00311-X
.svg)